What is Gene Expression? What is Lac Operon? Definition of Gene Regulation is “the control of gene regulation and expression is very well understood in prokaryotes. The switching on and off of the gene to regulate the synthesis of various proteins (the enzymes, for example) is essential. The fluctuation in the immediate environment of the organism is coped with by such regulation”. A group of genes that are regulated together is called “Operon“. This article gives the details of Lac Operon.
An E.Coli bacterial cell, for instance, is dependent for its nutrients on the whimsical eating habit of its human host in whose, but it is living.
If the bacterium does not get the amino acid tryptophan from the host’s food, it activates a pathway to make its own tryptophan (as it needs this amino acid in order to survive) from another compound.

The cell later stops the production of tryptophan if the human host eats the tryptophan-rich meal; this is how a cell avoids squandering its resources to produce a substance that is available from its surrounding in a prefabricated form. Like this, the bacteria tune their metabolism to changing environments.
What does the lac operon do? The basic mechanism for the control of gene expression was discovered in 1961 by Francois Jacob and Jacques Monod at the Pasteur Institute in Paris. Their findings became known as the Operon concept of Prokaryotic gene regulation. Why is it beneficial for organisms to control gene expression?

According to Wikipedia, The Operon means,
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be co-transcribed to define an operon.
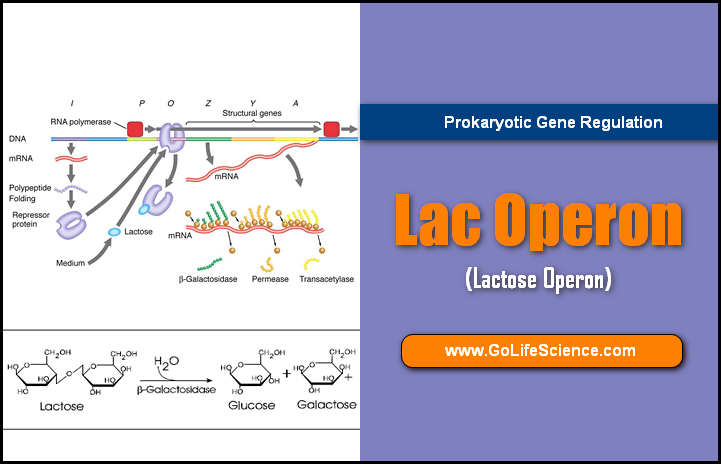
The Lac Operon
Jacob and Monod proposed the lac operon model to explain the regulation of the synthesis of an enzyme, β-galactosidase, the lac operon in e.coli, only when lactose is available as a substrate and Glucose is not available. This was the first study of metabolic regulation based on genetic analysis of the lac system. They called this the Operon Model (Lac Operon Concept).

- Structural Genes: Genes that code for proteins required by the cell (for functioning as an enzyme or to be structural proteins) are called Structural genes;
- Regulatory Genes: an overwhelming majority of the genes are in this category. Genes that code for proteins which, in turn, regulate the expression of other genes, are called Regulatory genes.
The crux of regulation is that a regulator gene codes for a regulator protein that controls transcription by binding to a particular site(s) on DNA. Regulation can be positive, when the interaction turns the gene on, or negative when the gene is turned off by the interaction.
The classic model of control of gene expression is negative; the most common mode of control in eukaryotes is positive. But, no type of regulation is exclusive to any one genome; there are examples of positive regulation in bacteria and negative regulation in eukaryotes.
How the Lac Operon Regulated?
How does lac operon work? Bacterial structural genes are mostly organized as clusters, consisting of genes coding for proteins whose functions are related. All the enzymes of a metabolic pathway are coded in a cluster of genes that are coordinately regulated. Simply it is a special gene machine.
There are three structural genes clustered together in Lac Operon. (An operon is a unit of bacterial or Prokaryotic gene expression and regulation, comprising structural genes and control elements (regulator genes), in DNA which is recognized by the regulatory proteins).
The genes are Lac Z, LacY, and Lac A. The gene products, the enzyme, enable the bacterial cells to take up and metabolize β-galactosidase, such as Lactose.

Lac Z :
The function of lacz gene is, which codes for the enzyme β-galactosidase; the enzyme breaks the β-galactoside into its components Sugar; E.g: Lactose is cleaved into Glucose and Galactose for further metabolism.

LacY:
Lac Y codes for the β-galactoside Permease; this transports the β-galactosides into the cell.

Lac A :
Lac A codes for β-galactoside transacetylase, an enzyme that transfers an acetyl group from acetyl~CoA to β-Galactosides.
The regulator genes for this cluster of three genes are the lac I gene, the lac O operator, and the LacP promoter. The entire system, comprising the structural genes and the regulatory (=control) elements, forms a common unit called the Lac Operon.
Prokaryotic Gene expression Steps

The products of the lacZ and LacY genes are encoded in a single polycistronic mRNA molecule. (The product of lacA gene is an enzyme (transacetylase) used in the metabolism of certain β-galactosides other than lactose. The positive and negative regulation of the lac operon is explaining below.
For the lac operon, the I-gene is regulator Gene, O is the Operator, and P is the Promoter to which RNA polymerase binds. The initiation site in the DNA, for transcription, is next to the operator and it is followed by the structural genes Z, Y, and A.
The three structural genes are transcribed to give a single mRNA molecule that codes for all three proteins. An mRNA molecule coding for more than one protein is known as a Polygenic (or Polycistronic) transcript.
The Lac operator has an asymmetric base sequence:
Walter Gilbert and his associates have sequenced the DNA region which comprises the operator. The base sequence is very interesting: 28 base pairs are related by a two-fold axis of symmetry.
Lac repressor protein binds to the operator and blocks transcription, in the absence of an inducer.
Walter Gilbert and Benno Muller-Hill showed that the lac repressor is a protein that binds to the lac operator DNA. It is a product of the LacI gene. It is a tetramer of 37kd subunits, each with a binding site for the inducer; the inducer is the lactose molecule. The Lac repressor has been shown to contain two-fold axes of symmetry, like the operator to which it binds.
When the repressor binds to the operator, its presence prevents RNA polymerase from initiating transcription at the promoter.
When is gene expression blocked in the lac operon system?
When E.coli cells are grown in the absence of lactose (or β-galactoside), there is no need for β -galactosidase and they have very few molecules of this less than 5 molecules per cell. In the absence of inducer (β-galactoside), the repressor molecules are produced by the LacI gene. The repressor prevents the transcription of the structural genes.
(i) Inducible System:
Active Repressor + Operator → System OFF
Active Repressor + Inducer = Inactive Repressor → System ON
(ii) Repressible System:
Apo-repressor and co-repressor complex = Active repressor → System OFF
Apo-repressor = Inactive Repressor → System ON
When a β-galactoside (like lactose) is added, the inducer inactivates the repressor and the latter cannot bind to the operator. RNA polymerase binds to the DNA (at the promoter-Operator site), and the structural genes are transcribed and the enzymes are produced. Within 2-3 minutes after the addition of the inducer, up to 500 molecules of β-galactosidase (enzyme) per bacterium are present.
If the lactose is removed from the medium, the synthesis of the enzyme stops as rapidly as it originally started. The repressor protein is the component that responds to the inducer. It has the role of controlling transcription of the component that responds to the inducer.
It has the role of controlling transcription of the lac ZYA structural genes in response to the environment. The start of the repressor determines whether the promoter is switched on or off:
- In the absence of the inducer, the genes are not transcribed; because the repressor protein is in an active form that is bound to the operator.
- When an inducer is added, the repressor is converted into an inactive form which leaves the operator, and then transcription starts at the promoter and proceeds through the genes to a terminator located somewhat beyond lacA.
In the absence of the inducer, the repressor is always bound to the operator (because of its high affinity) to ensure that the adjacent structural genes cannot be transcribed. It is to be remarked that
- The repressor can prevent transcription and
- The repressor can recognize the small molecule inducer.
The repressor has two binding sites, one for the inducer and one for the operator. When the inducer binds at its site, it changes the conformation of the repressor (protein) in such a way as to influence the activity of the other site. This type of relationship is an example of allosteric control.
Various modes of regulation:
Positive or negative control could be used to achieve either induction or repression by the appropriate interaction between the regulator protein and the small molecule inducer or co-repressor.

Induction is achieved when an inducer inactivates a repressor protein or activates an activator protein. Repression is accomplished when a co-repressor activates a repressor protein (Catabolite repression), to be under negative control. Genes that are active only when specifically turned on are said to be under positive control.
An operon system like the Lac operon in E.coli is rare or nonexistent in eukaryotes. Genetic crosses have revealed no operator genes and specific repressors have not been found.
Unlike the situation in prokaryotes, regulation in eukaryotes depends upon multiple mechanisms selectively control the rates of protein synthesis by different genes in different cells. Furthermore, these mechanisms operate at every stage in the formation and utilization of mRNA. Thus there are several levels of control.
- Differential transcription, perhaps the most significant category of control mechanisms
- Differential processing of primary RNA transcripts in the cell nucleus
- Differential translation of mRNA into protein
In prokaryotes, gene regulation has a limited role. The Lac operon (Lactose Operon) mechanism does permit sensitive responses to a rapidly changing environment.
However, ensuring adjustments are for the short term, briefly affecting cell division and growth without producing permanent alterations in Prokaryotic gene expression. This is in keeping with the short life cycles of these organisms.
Overview of the Lac Operon Gene Regulation

What happens to E.Coli when lactose is not present? The above lac operon concept represents the Lac operon concept in two conditions:
- This is the condition of absence of Lactose molecule in Prokaryotic gene expression
- This is the condition of the presence of Lactose molecule in Prokaryotic gene expression
