A fundamental property of living organisms is their ability to reproduce. Bacteria and fungi can divide to produce daughter cells that are identical to the parental cells. Sexually reproducing organisms produce offspring that are similar to themselves. This article covers the Enzymes Involved in DNA Replication.
On a cellular level, this reproduction occurs by mitosis, the process by which a single parental cell divides to produce two identical daughter cells.
- The Cell – Structure, and Functions (Synopsis Points)
- Structure of Cell Membrane: Basic Guide for Students
In the germline of sexually reproducing organisms, a parental cell with a diploid genome produces four germ cells with a haploid genome via a specialized process called “Meiosis“.
In both of these processes, the genetic material must be duplicated prior to cell division so that the daughter cells receive a full complement of the genetic information.
Thus accurate and complete replication of the DNA is essential to the ability of a cell organism to reproduce. This is the special and complete guide of Enzymes Involved in DNA Replication.
In the remaining sections of the chapter, we focus on the enzymes that mediate DNA replication. In these descriptions, you will encounter several cases of structure suggesting a particular function.
We will point out parallels and homologies between bacterial and eukaryotic DNA replication components.
This chapter covers the basic process and enzymology of DNA synthesis, and the next chapter will cover the regulation of DNA replication.
Replication means “Synthesis of daughter nucleic acid molecules identical to the parental nucleic acids”. In the replication, nucleic acids will be double by the Enzymes Involved in DNA Replication.

Table of Contents
Enzymes involved in DNA Replication
This is the list of dna replication enzymes and their functions. Let us discuss this in detail…
- Single-Stranded Binding Protein (SSBP)
- DNA Helicases
- Topoisomerases
- DNA primase
- DNA Ligase
- DNA polymerases
1. Single-Stranded Binding Protein (SSBP)
SSBP means Single-Stranded Binding Proteins. It has a very important role in DNA Replication in E.Coli.
Single-stranded binding proteins bind to and stabilize single-stranded DNA during DNA replication until the single-stranded DNA can be used as a template for a new strand to bind to.
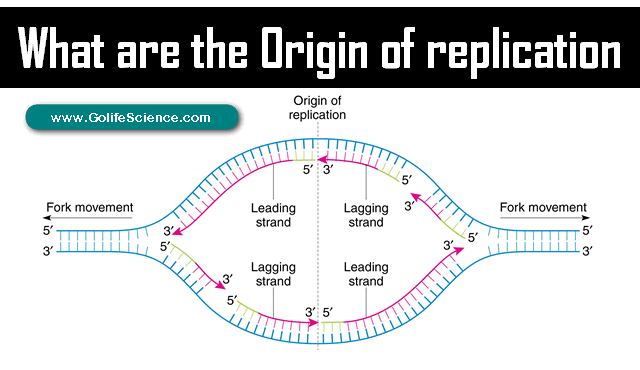
The SSB proteins are attached with both the lagging strand and the leading strand to prevent re-association of the strands.

- The molecular weight of the SSB protein is 75,600
- It contains FOUR identical subunits, which binds single-stranded DNA.
- The main function of the SSB protein is “Prevents reannealing” in the Replication process.
- As DNA Polymerase iii holoenzyme advances, it must displace the SSB protein in order that base pairing of the nucleotide being added can occur.
- “RFA” is a single-stranded DNA binding protein equivalent in function to the E.Coli SSB protein.
2.DNA Helicase
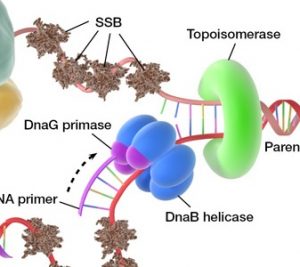
- DNA helicase enzyme functions “Unwinds DNA”.
- They have molecular weight 300,000, which contains SIX identical subunits.
- “Okazaki fragments” are short stretches of 1000-2000 bases produced during discontinuous replication, they are later joined into a covalently intact strand.
- The “Dna.B helicase” and “Dna.G Primase” constitute a functional unit within the replication complex, called the “PRIMOSOME”.
- The DNA is around by the Dna.B helicase at the replication fork, DNA primase occasionally associates with Dna.B helicase and synthesizes a short RNA primer.
- ”Helicase” and “Nuclease” activities of the Rec B, C, D enzyme is believed to help initiate homologous genetic recombination in E.Coli. It is also involved in the repair of double-strand breaks at the collapsed replication fork.
Function:
- A“Helicase” is an enzyme that separates the strands of DNA usually the hydrolysis of ATP to provide the necessary energy.
3. Topoisomerases
- Topoisomerase is also known as “DNA Gyrase”.
- “Topoisomerases” is an enzyme that can change the “Linking number”(Lk).
- Every cell has enzymes that increase (or) decrease the extent of DNA unwinding is called “Topoisomerases” the property of DNA that they change is the linking number.
- Topoisomerases”, these enzymes play an especially important role in processes such as “Replication” and “DNA packaging”.
- There are two classes of topoisomerases.
- Type-I Topoisomerases
- Type-II Topoisomerases
a) Type-I Topoisomerases:
This act by transiently breaking one of the two DNA strands, rotating one of the ends of the unbroken strand, and rejoining the broken ends; they change Lk in increments of 1.
b) Type-II Topoisomerases:
The enzyme breaks both DNA strands and changes Lk in increments of 2.
Prokaryotic Topoisomerases
FOUR different Topoisomerases (I and IV) occur in E.Coli.
1) Type.I (Topoisomerase I and III):
The type I generally relax DNA by removing negative super-coils (increasing Lk)
2) Type.II (Topoisomerase II and IV):
The Topoisomerase II is also called “DNA gyrase”, can introduce negative supercoils. (Decrease Lk).
- It uses the energy of ATP and a surprising mechanism to accomplish this.
- The degree of supercoiling of bacterial DNA is maintained by regulation of the net activity of topoisomerase-I and II.
Eukaryotic Topoisomerases
Eukaryotic cells also have type-I and type-II topoisomerases. Topoisomerases-I & II are both type-I. The two type-II topoisomerases, topoisomerases IIa and IIb, can not unwind DNA (introduce negative supercoils).
Although both can relax positive and negative supercoils. We consider one probable origin of negative supercoils in eukaryotic cells.
- The DNA gyrase molecular weight is 400,000, which contains FOUR subunits and functions “Supercoiling”.
- Supercoiled DNA is a higher-ordered structure occurring in circular DNA molecules wrapped around a core.
What is the linking number? The linking number (Lk) is a topological property. Lk can be defined as “ the number of times the second strand pierces the second strand surface”.
4. DNA Primase
In replication, before DNA polymerase iii can begin synthesizing DNA primers must be present on the template generally short segments of RNA synthesized by an enzyme called “Primases”.
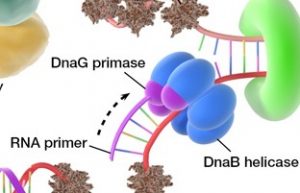
- DNA primase has molecular weight 60,000 Dalton and contains only a single subunit, which functions synthesize RNA primers.
- The Dna.B helicase and Dna.G primase constitute a functional unit within the replication complex, called the “Primosome”.
- The RNA primer typically is 15-50 bases long. It synthesizes primers starting with the sequence pppAG, opposite the sequence 3’-GTC-5’ in the template.
5. DNA Ligase
An enzyme that creates a phosphodiester bond between the 3’ end of one DNA segment and the 5’ end of another. Once the RNA primer has been removed and replaced the adjacent Okazaki fragments must be linked together. The 3’-OH end of one fragment is adjacent to the 5’-Phosphate end of the previous fragment. The responsible for sealing this nick lies with the enzyme DNA ligase. Ligases are present in both prokaryotes and eukaryotes.
Mechanism of Enzyme activity:
The E.Coli and T4 ligases share the property of sealing nicks that have 3’’-OH and 5’- P termini. Both enzymes undertake a two-step reaction, involving an ‘enzyme-AMP complex’.
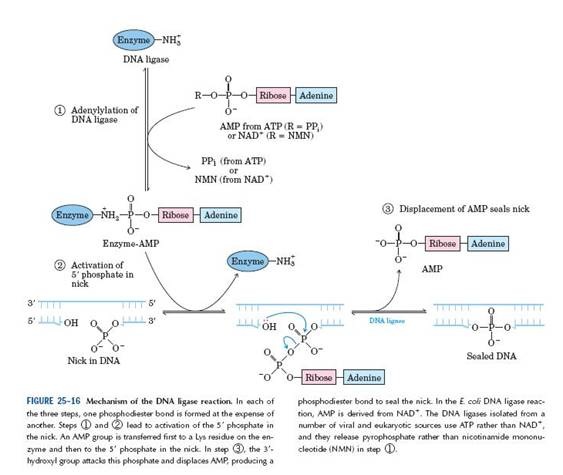
- The E.Coli and T4 enzyme use different cofactors.
- The E.Coli enzymes use NAD as a cofactor, the T4 enzyme uses ATP.
The AMP of the enzyme complex becomes attached to the 5’-Phosphate of the nick; and then a phosphodiester bond is formed with the 3’-OH terminus of the nick, releasing the enzyme and the AMP.
Final words
In this chapter (DNA replication enzymes and their functions) and the next, we will examine the process of replication.
After describing the basic mechanism of DNA replication, we discuss the various techniques researchers have used to achieve a complete understanding of replication.
Indeed, a theme of this chapter is the combination of genetic and biochemical approaches that have allowed us to uncover the mechanism and physiology of DNA replication.