Repetitive DNA is one of the most intriguing and complex components of the genome. It plays a vital role in genome organization, genetic variation, and evolutionary genetics. Despite its prevalence, repetitive DNA is often misunderstood and dismissed as “junk DNA.” However, modern research has revealed that these sequences are far from useless.
This article will explore what repetitive DNA, its types, functions, and significance in genome stability and genetic variation. Whether you’re a student or a teacher, this guide will provide you with a clear understanding of repetitive DNA and its importance in life sciences.
Table of Contents
What is repetitive DNA?
Repetitive DNA refers to sequences of DNA that are repeated multiple times throughout the genome. These sequences can range from a few base pairs to several thousand base pairs in length. Repetitive DNA sequences are a significant component of the genome, especially in eukaryotes, where they can constitute a large portion of the DNA.
The repetitive DNA definition can be summarized as DNA sequences that are present in multiple copies within the genome. These sequences can be either tandem repeats, where the repeats are adjacent to each other, or dispersed repeats, where the repeats are scattered throughout the genome.
In contrast to unique DNA, which consists of sequences that appear only once in the genome, repetitive DNA is characterized by its sequence repetition. While unique DNA codes for proteins and functional RNA, repetitive DNA is often found in non-coding DNA regions, although it can also be present within genes.
Types of Repetitive DNA
There are several types of repetitive DNA sequences, each with its own characteristics and functions. These can be broadly categorized into tandem repeats and dispersed repeats.
1. Tandem Repeats
Tandem repeats are sequences that are repeated one after another in a head-to-tail fashion. These repeats can be further classified into satellite DNA, minisatellite DNA, and microsatellite DNA.
a. Satellite DNA
Satellite DNA consists of large arrays of tandemly repeated sequences, often found in the centromeric and telomeric regions of chromosomes. These sequences are typically highly repetitive DNA and can span millions of base pairs.
b. Minisatellite DNA
Minisatellite DNA consists of moderately sized tandem repeats, usually ranging from 10 to 60 base pairs in length. These sequences are often found in the subtelomeric regions of chromosomes and are known for their role in genetic variation.
c. Microsatellite DNA
Microsatellite DNA, also known as short tandem repeats (STRs), consists of very short sequences, usually 2-6 base pairs in length, repeated in tandem. These sequences are highly polymorphic and are widely used in genetic fingerprinting and evolutionary genetics.
2. Dispersed Repeats
Dispersed repeats are sequences that are scattered throughout the genome rather than being clustered together. These repeats include transposable elements, which are capable of moving within the genome, and other repetitive elements that do not move.
Transposable Elements
Transposable elements, also known as “jumping genes,” are sequences that can change their position within the genome. They play a significant role in genome evolution and can contribute to genetic variation and genome stability.
Single Copy vs Repetitive DNA
The genome is composed of both single copy DNA and repetitive DNA. Single copy DNA includes genes that code for proteins and functional RNA, while repetitive DNA consists of sequences that are repeated multiple times. The balance between these two types of DNA is crucial for genome organization and function.
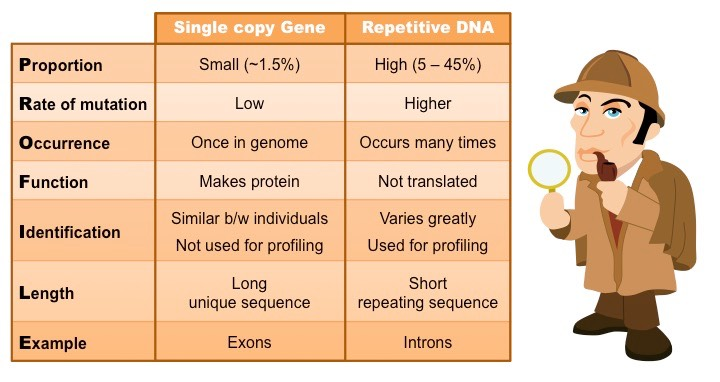
a. Single Copy DNA
Single copy DNA is unique within the genome and typically codes for proteins and functional RNA. These sequences are essential for the normal functioning of the organism and are highly conserved across species.
b. Repetitive DNA
Repetitive DNA, on the other hand, is characterized by its sequence repetition. While some repetitive sequences have known functions, others are still poorly understood. However, it is clear that repetitive DNA plays a crucial role in genome stability, genetic variation, and evolutionary genetics.
Repetitive DNA in Eukaryotes
Repetitive DNA is particularly abundant in eukaryotes, where it can constitute a significant portion of the genome. In humans, for example, repetitive DNA makes up approximately 50% of the genome. This high prevalence suggests that repetitive DNA plays an important role in the biology of eukaryotic organisms.
a. Highly Repetitive DNA
Highly repetitive DNA consists of sequences that are repeated thousands or even millions of times within the genome. These sequences are often found in the centromeric and telomeric regions of chromosomes and are thought to play a role in chromosome structure and genome stability.
b. Moderately Repetitive DNA
Moderately repetitive DNA consists of sequences that are repeated hundreds to thousands of times within the genome. These sequences can be found throughout the genome and include transposable elements and other repetitive elements.
Role of Repetitive DNA
Despite being often labeled as “junk DNA,” repetitive DNA has several important functions within the genome. These functions include genome stability, genetic variation, and evolutionary genetics.
a. Genome Stability
Repetitive DNA plays a crucial role in maintaining genome stability. For example, satellite DNA in the centromeric regions of chromosomes is essential for proper chromosome segregation during cell division. Similarly, telomeric repeats protect the ends of chromosomes from degradation and fusion.
b. Genetic Variation
Repetitive DNA is a major source of genetic variation. Minisatellite and microsatellite DNA are highly polymorphic, meaning that they vary greatly between individuals. This variation is used in genetic fingerprinting and can also contribute to evolutionary genetics.
c. Evolutionary Genetics
Repetitive DNA plays a significant role in evolutionary genetics. Transposable elements, for example, can cause mutations and rearrangements within the genome, leading to genetic variation and potentially driving evolution. Additionally, the expansion and contraction of tandem repeats can contribute to genome evolution.
Repetitive DNA and Junk DNA
The term “junk DNA” was originally used to describe non-coding DNA, including repetitive DNA, that was thought to have no function. However, it is now clear that much of this DNA plays important roles in genome organization, genetic variation, and evolutionary genetics. While some repetitive DNA may still be nonfunctional, it is increasingly recognized that these sequences are far from being “junk.”
Advanced Insights into Repetitive DNA
1. Discovery of Repetitive DNA
The discovery of repetitive DNA was made possible through the technique of DNA denaturation and renaturation. When genomic DNA is denatured (made single-stranded) and then allowed to renature, the rate at which it does so can reveal the presence of repetitive sequences. Repetitive DNA renatures more quickly because multiple copies allow a strand to find a partner more easily, whereas unique DNA renatures more slowly.
2. Processed Pseudogenes
In addition to SINEs and LINEs, which constitute the bulk of the moderately repeated DNA in mammals, other moderately repetitive sequences have been identified. Many of these represent mutated DNA copies of a wide variety of mRNAs that have integrated into chromosomal DNA.
These are not duplicates of whole genes that have drifted into functionality (i.e., the pseudogenes discussed earlier in this chapter) because they lack introns and do not have flanking sequences similar to those of the functional gene copies.
Instead, these DNA segments appear to be retrotransposed copies of spliced and polyadenylated (processed) mRNA. Compared with normal genes encoding mRNAs, these inserted segments generally contain multiple mutations, which are thought to have accumulated since their mRNAs were first reverse transcribed and randomly integrated into the genome of a germ cell in an ancient ancestor.
These nonfunctional genomic copies of mRNAs are referred to as processed pseudogenes. Most processed pseudogenes are flanked by short direct repeats, supporting the hypothesis that they were generated by rare retrotransposition events involving cellular mRNAs.
3. Retrotransposition of Small RNAs
Other moderately repetitive sequences representing partial or mutant copies of genes encoding small nuclear RNAs (snRNAs) and tRNAs are found in mammalian genomes. Like processed pseudogenes, these nonfunctional copies of small RNA genes are flanked by short direct repeats and most likely result from rare retrotransposition events that have accumulated through the course of evolution.
Enzymes expressed from a LINE or viral retrotransposon are thought to have carried out the retrotransposition of mRNAs, snRNAs, and tRNAs.
4. Gene Conversion and Repetitive DNA
The function of repetitive DNA was illuminated by several lines of genome research during the 1980s. Strong evidence was presented showing that repetitive DNA is an evolutionary device to catalyze the formation of new genes by suppressing gene conversion. This evidence has been gathered here so that a new generation of genomic scientists may examine it in the fresh light of reason.
Gene conversion links similar DNA sequences together. It can operate on genes within a multigene family, or it can operate interchromosomally on gene homologs. Similar DNA sequences are the substrates for gene conversion. Gene conversion is the cohesive force allowing species to exist. The gene pool of a species consists of DNA sequences in a network linked by gene conversion events.
Repetitive sequences play the role of uncoupling this network, thereby allowing new genes to evolve. The shorter Alu or SINE repetitive DNA are specialized for uncoupling intra-chromosomal gene conversion, while the longer LINE repetitive DNA are specialized for uncoupling interchromosomal gene conversion.
5. Repetitive DNA and Genome Function
There are clear theoretical reasons and many well-documented examples that show that repetitive DNA is essential for genome function. Generic repeated signals in the DNA are necessary to format the expression of unique coding sequence files and to organize additional functions essential for genome replication and accurate transmission to progeny cells.
Repetitive DNA sequence elements are also fundamental to the cooperative molecular interactions forming nucleoprotein complexes. Here, we review the surprising abundance of repetitive DNA in many genomes, describe its structural diversity, and discuss dozens of cases where the functional importance of repetitive elements has been studied in molecular detail.
Repetitive DNA and Disease
Satellite DNA, also known as tandemly repeated DNA, consists of clusters of repeated sequences and represents a diverse class of highly repetitive elements. Satellite DNA can be divided into several classes according to the size of an individual repeat: microsatellites, minisatellites, minisatellites, and microsatellites. Originally considered as “junk” DNA, satellite DNA has more recently been reconsidered as having various functions.
Moreover, due to the repetitive nature of the composing elements, their presence in the genome is associated with high-frequency mutations, epigenetic changes, and modifications in gene expression patterns, with a potential to lead to human disease.
Therefore, the satellite DNA study will be beneficial for developing a treatment of satellite-related diseases, such as FSHD, neurological and developmental disorders, and cancers.
a. Clinical Relevance of Satellite DNA
In recent years, much attention has been brought to the role of repeat sequences in various pathologies, such as epilepsy, embryonic lethality, and cancers. The increase in the number of copies of repeats in genomic DNA is the single most important cause of nearly 30 hereditary disorders:
- X-fragile syndrome
- Myotonic dystrophy
- Huntington’s disease
- Friedreich’s ataxia
Scott et al. have described the first evidence of the insertion of 18 beta-satellite units into the gene coding transmembrane serine protease, resulting in autosomal recessive deafness.
b. Cancer as a Satellite-Related Disease
“Breaking of satellites’ silence” is now a new paradigm in cancerogenesis. While gene-specific loci can be either hypo- or hypermethylated in cancer, the highly repeated DNA sequences are only hypomethylated in this disease. Moreover, the global DNA hypomethylation observed so frequently in cancers is mostly due to satellite DNA.
Ehrlich et al. were the first to demonstrate the hypomethylation of tandem minisatellite centromeric DNA (namely centromere-adjacent satellite 2, (Sat2)) in breast adenocarcinomas, ovarian epithelial tumors, and Wilms tumors. A highly significant difference in the methylation level was found in satellite repeats (SATR1 and ARLa) in neurosarcoma.
In 76% of hepatocellular carcinoma cases, the reduction in Sat2 methylation has also been demonstrated. More recently, Zhu et al. showed that the loss of the BRCA1 tumor suppressor gene provoked satellite-DNA derepression in breast and ovarian tumors of mice and humans.
| Type of Repetitive DNA | Description | Examples |
|---|---|---|
| Satellite DNA | Large arrays of tandemly repeated sequences, often found in centromeric and telomeric regions. | Centromeric repeats, Telomeric repeats |
| Minisatellite DNA | Moderately sized tandem repeats, usually 10-60 base pairs in length. | VNTRs (Variable Number Tandem Repeats) |
| Microsatellite DNA | Very short tandem repeats, usually 2-6 base pairs in length. | STRs (Short Tandem Repeats) |
| Transposable Elements | Sequences that can change their position within the genome. | LINEs (Long Interspersed Nuclear Elements), SINEs (Short Interspersed Nuclear Elements) |
| Function | Description | Examples |
|---|---|---|
| Genome Stability | Maintains the structural integrity of chromosomes. | Satellite DNA in centromeres, Telomeric repeats |
| Genetic Variation | Provides a source of variation between individuals. | Minisatellites, Microsatellites |
| Evolutionary Genetics | Contributes to genome evolution and adaptation. | Transposable elements, Tandem repeats |
Conclusion
Repetitive DNA is a complex and dynamic component of the genome that plays a crucial role in genome organization, genetic variation, and evolutionary genetics. Despite being often dismissed as “junk DNA,” these sequences have important functions that are essential for the stability and evolution of the genome.
By understanding the types of repetitive DNA and their roles, students and teachers can gain a deeper appreciation for the complexity and beauty of the genome.