Enzymes are the workhorses of the cell, catalyzing biochemical reactions with remarkable specificity and efficiency. However, without proper regulation, these powerful catalysts could lead to chaos within the cell. Enzyme regulation is a critical aspect of cellular metabolism, ensuring that biochemical pathways operate smoothly and respond appropriately to the cell’s needs. In this article, we will explore the mechanisms of enzyme regulation, their importance in maintaining cellular homeostasis, and their implications in health and disease.
Table of Contents
Features of Enzyme Regulation
Enzyme regulation is a crucial process in living organisms that controls the rate of enzymatic reactions. This regulation ensures that metabolic pathways operate efficiently and respond appropriately to changing cellular conditions. By modulating enzyme activity, cells can maintain homeostasis, conserve energy, and adapt to environmental changes.
Enzyme regulation is characterized by several key features that contribute to its effectiveness and versatility in controlling cellular processes:
- Specificity: Enzyme regulation mechanisms are highly specific, targeting particular enzymes or enzyme families. This specificity ensures that control is exerted precisely where it’s needed without affecting unrelated metabolic pathways.
- Rapid response: Many regulatory mechanisms can quickly activate or inhibit enzymes in response to cellular signals. This rapid response allows organisms to adapt swiftly to changing conditions.
- Reversibility: Most forms of enzyme regulation are reversible, allowing for dynamic control of enzyme activity. This reversibility enables fine-tuning of metabolic processes and helps maintain cellular homeostasis.
- Multi-level control: Enzyme regulation occurs at multiple levels, including genetic, transcriptional, translational, and post-translational stages. This multi-tiered approach provides comprehensive control over enzyme activity.
- Integration with signalling pathways: Enzyme regulation is often integrated with cellular signalling cascades, allowing for coordinated responses to internal and external stimuli.
- Amplification: Some regulatory mechanisms, particularly those involving allosteric enzymes, can produce amplified effects. A small change in effector concentration can result in a significant change in enzyme activity.
- Energy efficiency: Many regulatory mechanisms, such as allosteric regulation, do not require additional energy input, making them energetically efficient ways to control metabolism.
- Feedback control: Enzyme regulation often involves feedback loops, where the products of a metabolic pathway can influence the activity of enzymes earlier in the pathway. This feature helps maintain metabolic balance and prevent the overproduction of metabolites.
- Cooperativity: Some regulatory mechanisms, like those seen in haemoglobin, exhibit cooperativity, where the binding of one substrate molecule influences the binding of subsequent molecules. This phenomenon allows for sharp, switch-like responses to changes in substrate concentration.
- Compartmentalization: Enzyme regulation can be influenced by the spatial organization of enzymes within cells. Compartmentalization in organelles or multi-enzyme complexes can enhance the efficiency and specificity of regulation.
These features work in concert to create a flexible, responsive, and precise system for controlling enzyme activity. Understanding these characteristics is crucial for appreciating the sophistication of enzyme regulation and its role in maintaining cellular function.
Enzymes and Their Role in Metabolism
Enzymes are biological catalysts that accelerate chemical reactions by lowering the activation energy required for the reaction to proceed. They are typically proteins, although some RNA molecules, known as ribozymes, also exhibit catalytic activity. Enzymes are involved in virtually every biochemical process in the cell, from DNA replication and protein synthesis to energy production and waste elimination.
Metabolism, the sum of all chemical reactions in the cell, is divided into two main categories: catabolism and anabolism. Catabolic pathways break down complex molecules into simpler ones, releasing energy in the process. Anabolic pathways, on the other hand, use energy to build complex molecules from simpler ones. Enzymes play a central role in both catabolic and anabolic pathways, ensuring that these reactions occur at rates compatible with life.
Need for Enzyme Regulation
Given the central role of enzymes in metabolism, it is crucial that their activity is tightly regulated. Unregulated enzyme activity could lead to the overproduction or underproduction of metabolites, disrupting cellular homeostasis. For example, excessive activity of enzymes involved in glucose metabolism could lead to hyperglycemia, while insufficient activity could result in hypoglycemia.
Enzyme regulation allows cells to respond to changes in their internal and external environments. For instance, when glucose levels are high, cells can upregulate enzymes involved in glycolysis to convert glucose into energy. Conversely, when glucose levels are low, cells can downregulate these enzymes and upregulate those involved in gluconeogenesis to produce glucose from non-carbohydrate sources.
Types of Enzyme Regulation
Enzyme regulation encompasses a wide array of mechanisms that operate at different levels of cellular organization. These regulatory strategies can be broadly categorized into several types:
1. Allosteric Regulation
Allosteric regulation is a common mechanism of enzyme control in which the binding of a molecule (an allosteric effector) at a site other than the enzyme’s active site alters the enzyme’s activity. Allosteric effectors can be either activators or inhibitors. Activators increase enzyme activity, while inhibitors decrease it.
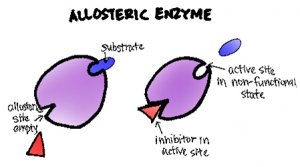
Allosteric regulation can be either positive (activation) or negative (inhibition). Positive allosteric modulators enhance enzyme activity, while negative modulators decrease it. A classic example of allosteric regulation is seen in phosphofructokinase-1 (PFK-1), a key enzyme in glycolysis. ATP acts as a negative allosteric regulator of PFK-1, while AMP acts as a positive regulator, allowing the enzyme to respond to the cell’s energy status.
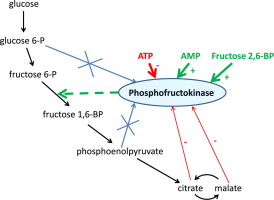
The beauty of allosteric regulation lies in its ability to provide rapid, reversible control without the need for covalent modifications or changes in enzyme concentration. This makes it an energetically efficient and responsive mechanism for fine-tuning enzyme activity.
Allosteric regulation is often used to control metabolic pathways. For example, the enzyme phosphofructokinase-1 (PFK-1), which catalyzes a key step in glycolysis, is allosterically regulated by ATP and AMP. When ATP levels are high, ATP binds to an allosteric site on PFK-1, inhibiting its activity and slowing down glycolysis. Conversely, when ATP levels are low and AMP levels are high, AMP binds to PFK-1, activating it and stimulating glycolysis.
Allosteric regulation is a key mechanism in enzyme control. It occurs when a molecule binds to a site on the enzyme other than the active site, causing a change in the enzyme’s shape and activity. This binding site is called the allosteric site.
There are two main types of allosteric regulation:
- Positive allosteric modulation: An effector molecule enhances the enzyme’s activity.
- Negative allosteric modulation: An effector molecule decreases the enzyme’s activity.
Allosteric regulators can be classified as:
- Homotropic: The substrate itself acts as the regulator.
- Heterotropic: A molecule different from the substrate acts as the regulator.
Two models explain allosteric regulation:
- Concerted (MWC) model: All enzyme subunits change conformation simultaneously.
- Sequential (KNF) model: Subunits change conformation individually as they bind to the substrate.
Allosteric regulation plays a crucial role in many biological processes, including oxygen binding in hemoglobin and control of metabolic pathways like glycolysis.
2. Covalent Modification
Covalent modification involves the addition or removal of chemical groups to specific amino acid residues on the enzyme. This type of regulation can dramatically alter an enzyme’s activity, stability, or interactions with other molecules.
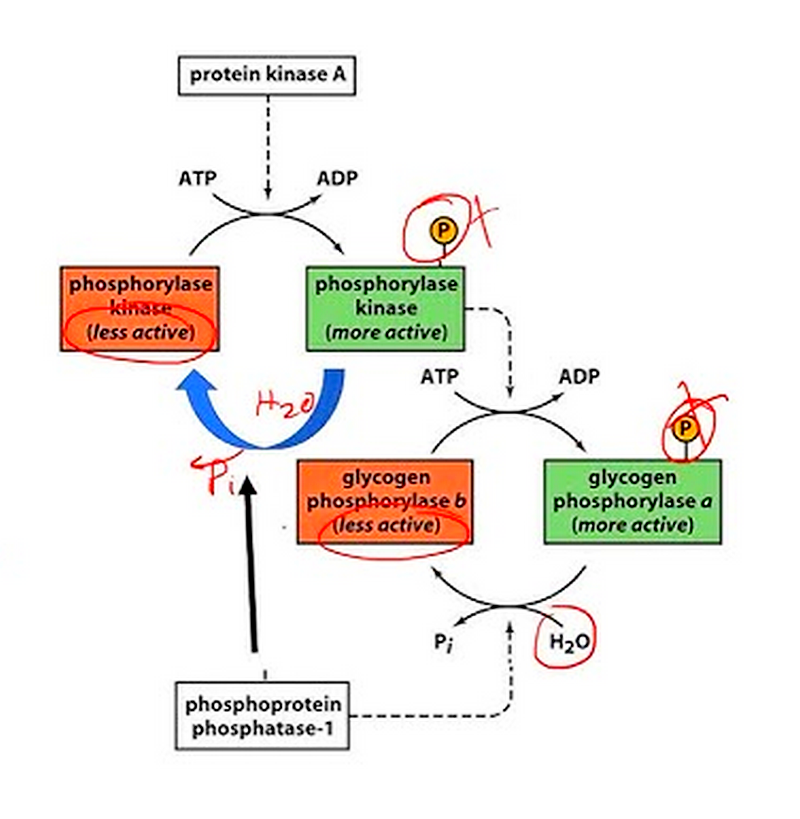
Common covalent modifications include:
a. Phosphorylation:
The addition of phosphate groups, often catalyzed by protein kinases. This is one of the most common and well-studied modifications.
Enzyme + ATP → Enzyme-PO₃²⁻ + ADP
Example: In glycogen metabolism, glycogen phosphorylase (GP) is activated by phosphorylation:
GP (inactive) + ATP → GP-PO₃²⁻ (active) + ADP
b. Acetylation:
The addition of acetyl groups, typically to lysine residues.
Enzyme + Acetyl-CoA → Enzyme-COCH₃ + CoA
Example: Histone acetylation in gene regulation:
Histone + Acetyl-CoA → Histone-COCH₃ + CoA
c. Methylation:
The addition of methyl groups, often to lysine or arginine residues.
Enzyme + S-Adenosyl methionine (SAM) → Enzyme-CH₃ + S-Adenosyl homocysteine (SAH)
Example: DNA methyltransferase (DNMT) in epigenetic regulation:
DNA + SAM → DNA-CH₃ + SAH
d. Ubiquitination:
The addition of ubiquitin molecules, often targeting proteins for degradation. This process involves a cascade of enzymes (E1, E2, E3).
Enzyme + Ubiquitin + ATP → Enzyme-Ubiquitin + AMP + PPi
Example: Degradation of cyclins in cell cycle regulation:
Cyclin + Ubiquitin + ATP → Cyclin-Ubiquitin + AMP + PPi
Phosphorylation is particularly prevalent in enzyme regulation. For example, glycogen phosphorylase, an enzyme involved in glycogen breakdown, is activated by phosphorylation in response to hormonal signals. This allows for rapid mobilization of glucose reserves when energy demands increase.
Covalent modifications provide a versatile means of regulation, as they can be reversible (through the action of enzymes that remove the modifications) and can integrate multiple signalling pathways to fine-tune enzyme activity.
3. Transcriptional Regulation
Transcriptional regulation controls enzyme production at the gene level by modulating mRNA synthesis. By modulating the rate of mRNA synthesis, cells can adjust the amount of enzyme available over longer time scales. This type of regulation involves:
a. Transcription factors (TFs):
Proteins that bind to specific DNA sequences to enhance or repress gene transcription.
TF + DNA ⇌ TF-DNA complex
Example: The lac repressor in E. coli: LacI (repressor) + lac operator ⇌ LacI-operator complex
When the repressor is bound, transcription is inhibited:
RNA polymerase + Promoter + LacI-operator complex → No transcription
When lactose is present, it binds to the repressor, causing it to release from the DNA: LacI-operator complex + Lactose → LacI-Lactose + Free operator RNA polymerase + Free promoter → mRNA transcription
b. Promoter regions:
DNA sequences that control the initiation of transcription.
RNA polymerase + Promoter ⇌ RNA polymerase-Promoter complex
Example: The TATA box in eukaryotes: TBP (TATA-binding protein) + TATA box ⇌ TBP-TATA complex RNA polymerase II + TBP-TATA complex → Transcription initiation
c. Enhancers and silencers:
Regulatory elements that can increase or decrease transcription rates.
Enhancer:
TF + Enhancer → TF-Enhancer complex
TF-Enhancer complex + RNA polymerase-Promoter complex → Increased transcription
Silencer:
Repressor + Silencer → Repressor-Silencer complex
Repressor-Silencer complex + RNA polymerase-Promoter complex → Decreased transcription
Example: The β-globin locus control region (LCR) in humans: LCR-binding factors + LCR → LCR-protein complex LCR-protein complex + β-globin promoter → Enhanced β-globin transcription
These mechanisms work together to fine-tune gene expression:
- Basal transcription: RNA polymerase + Promoter → Low-level mRNA production
- Activated transcription: TF + Enhancer + RNA polymerase + Promoter → High-level mRNA production
- Repressed transcription: Repressor + Silencer + RNA polymerase + Promoter → Minimal or no mRNA production
The interplay of these elements allows for precise control of enzyme levels in response to cellular needs and environmental signals. This regulation can lead to long-term adaptations and plays a crucial role in developmental processes and tissue-specific enzyme expression.
An example of transcriptional regulation is the lac operon in E. coli, which controls the production of enzymes necessary for lactose metabolism. In the absence of lactose, a repressor protein binds to the operator region, preventing transcription. When lactose is present, it binds to the repressor, causing it to release from the DNA and allowing transcription to proceed.
Transcriptional regulation allows for long-term adaptation to environmental conditions and plays a crucial role in developmental processes and tissue-specific enzyme expression.
4. Translational Regulation
Translational regulation controls the synthesis of enzymes at the mRNA level. This can involve:
- mRNA stability: Factors that influence the half-life of mRNA molecules
- Translational efficiency: Mechanisms that affect how efficiently ribosomes translate mRNA into protein
- RNA interference: Small RNA molecules that can inhibit translation or promote mRNA degradation
For instance, iron-responsive elements (IREs) in the mRNA of ferritin, an iron storage protein, allow for translational regulation based on iron availability. When iron levels are low, iron regulatory proteins bind to the IREs, inhibiting translation. When iron is abundant, the regulatory proteins are released, allowing translation to proceed.
Translational regulation provides an additional layer of control over enzyme production, allowing for rapid adjustments in enzyme levels without the need for changes in transcription.
5. Compartmentalization
Spatial organization within cells plays a crucial role in enzyme regulation. By compartmentalizing enzymes in specific organelles or creating multi-enzyme complexes, cells can control:
- Substrate availability: Limiting access to substrates can regulate enzyme activity
- Local concentrations: Creating microenvironments with high concentrations of enzymes and substrates can enhance reaction rates
- Channelling: Direct transfer of intermediates between enzymes in a pathway, increasing efficiency and reducing side reactions
An example of compartmentalization is the citric acid cycle, which occurs in the mitochondrial matrix. This localization ensures that the enzymes have access to the necessary substrates and cofactors while separating the process from potentially interfering cytosolic reactions.
6. Feedback Inhibition
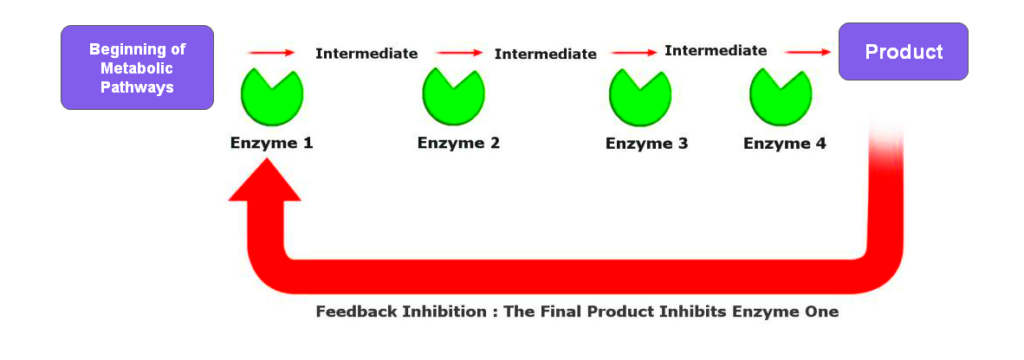
Feedback inhibition is a regulatory mechanism where the end product of a metabolic pathway inhibits an enzyme early in that pathway. It creates a negative feedback loop that helps maintain metabolic balance and prevent the overproduction of metabolites.
A classic example is the regulation of the amino acid biosynthesis pathway. In the production of isoleucine, the end product inhibits the first enzyme in the pathway, threonine deaminase. It ensures that isoleucine is only produced when needed and conserves energy and resources when isoleucine is abundant.
A well-known example of feedback inhibition is the regulation of the amino acid synthesis pathway. In bacteria, the end product of the pathway, such as isoleucine, can bind to and inhibit the first enzyme in the pathway, threonine deaminase. This prevents the overproduction of isoleucine and ensures that resources are not wasted on unnecessary synthesis.
Feedback inhibition allows for rapid and efficient control of metabolic flux, responding dynamically to the cell’s needs.
7. Zymogen Activation
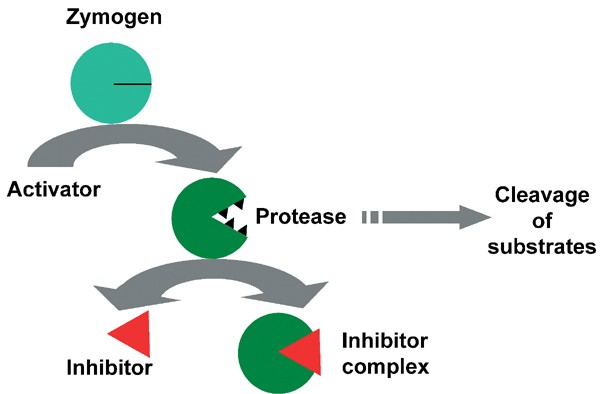
Some enzymes are synthesized as inactive precursors called zymogens. These zymogens require specific conditions or proteolytic cleavage to become active. This mechanism provides a powerful means of regulating potentially harmful enzymes, such as digestive enzymes and those involved in blood clotting.
For example, pepsinogen, the zymogen form of the digestive enzyme pepsin, is activated by the acidic environment of the stomach. This ensures that the protein-degrading activity of pepsin is only unleashed in the appropriate location, protecting the cells that produce and store the enzyme.
Zymogen activation allows for the spatial and temporal control of enzyme activity, preventing premature or inappropriate enzyme action.
8. Isozyme Production
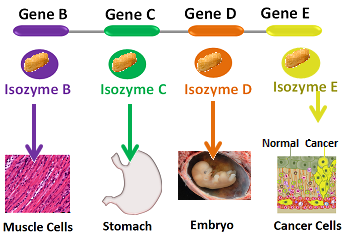
Isozymes are different forms of enzymes that catalyze the same reaction but have different kinetic or regulatory properties. By producing different isozymes, organisms can fine-tune enzyme activity for specific tissues or conditions.
Lactate dehydrogenase (LDH) is an example of an enzyme with multiple isozymes. The heart and skeletal muscle express different LDH isozymes optimized for their specific metabolic needs. The muscle isozyme favors the conversion of pyruvate to lactate during intense exercise, while the heart isozyme favors the reverse reaction, allowing for lactate utilization as a fuel source.
Isozyme production provides a means of tissue-specific regulation and adaptation to different physiological demands.
Enzyme Regulation in Metabolic Pathways
Enzyme regulation is crucial for the proper functioning of metabolic pathways. Below, we discuss how enzyme regulation operates in some key metabolic pathways.
1. Glycolysis and Gluconeogenesis
Glycolysis, the breakdown of glucose to produce energy, and gluconeogenesis, the synthesis of glucose from non-carbohydrate sources, are tightly regulated to ensure that they do not occur simultaneously. Key enzymes in these pathways are regulated by allosteric effectors and covalent modification.
For example, the enzyme pyruvate kinase, which catalyzes the final step of glycolysis, is inhibited by ATP and alanine, signaling that the cell has sufficient energy and building blocks. Conversely, it is activated by fructose-1,6-bisphosphate, indicating that glycolysis is actively producing energy.
In gluconeogenesis, the enzyme fructose-1,6-bisphosphatase is inhibited by AMP and activated by ATP, ensuring that glucose synthesis occurs only when energy levels are low.
2. Citric Acid Cycle
The citric acid cycle, also known as the Krebs cycle, is a central metabolic pathway that generates energy through the oxidation of acetyl-CoA. Key enzymes in the cycle are regulated by the availability of substrates and the energy status of the cell.
For example, the enzyme isocitrate dehydrogenase is activated by ADP, indicating a need for energy, and inhibited by ATP and NADH, signaling that the cell has sufficient energy and reducing equivalents.
3. Fatty Acid Metabolism
Fatty acid metabolism involves both the synthesis and breakdown of fatty acids, which are regulated by different enzymes. The enzyme acetyl-CoA carboxylase, which catalyzes the first step in fatty acid synthesis, is activated by citrate and inhibited by palmitoyl-CoA, ensuring that fatty acid synthesis occurs only when there is an abundance of acetyl-CoA and a need for fatty acids.
Conversely, the enzyme carnitine palmitoyltransferase I, which is involved in fatty acid oxidation, is inhibited by malonyl-CoA, a product of fatty acid synthesis. This prevents the simultaneous synthesis and breakdown of fatty acids.
Enzyme Regulation in Disease
Dysregulation of enzyme activity can lead to a variety of diseases. Below, we discuss some examples of how enzyme regulation is implicated in disease.
1. Cancer
Cancer is often associated with the dysregulation of enzymes involved in cell growth and division. For example, the enzyme telomerase, which maintains the ends of chromosomes, is typically inactive in most adult cells. However, in cancer cells, telomerase is often reactivated, allowing the cells to divide indefinitely.
Similarly, mutations in the genes encoding enzymes involved in the regulation of the cell cycle, such as cyclin-dependent kinases (CDKs), can lead to uncontrolled cell proliferation.
2. Metabolic Disorders
Metabolic disorders, such as diabetes and obesity, are often associated with the dysregulation of enzymes involved in glucose and lipid metabolism. For example, in type 2 diabetes, the enzyme glucokinase, which is involved in glucose sensing in the pancreas, may be less responsive to changes in blood glucose levels, leading to impaired insulin secretion.
In obesity, the enzyme lipoprotein lipase, which is involved in the breakdown of triglycerides in the blood, may be overactive, leading to the excessive storage of fat.
3. Neurological Disorders
Neurological disorders, such as Alzheimer’s disease and Parkinson’s disease, are often associated with the dysregulation of enzymes involved in the metabolism of neurotransmitters. For example, in Alzheimer’s disease, the enzyme acetylcholinesterase, which breaks down the neurotransmitter acetylcholine, may be overactive, leading to a deficiency of acetylcholine and impaired cognitive function.
In Parkinson’s disease, the enzyme monoamine oxidase B, which breaks down the neurotransmitter dopamine, may be overactive, leading to a deficiency of dopamine and impaired motor function.
Pharmacological Targeting of Enzyme Regulation
Given the central role of enzymes in disease, they are often targeted by drugs. For example, many cancer drugs inhibit enzymes involved in cell division, such as CDKs and telomerase. Similarly, drugs used to treat metabolic disorders often target enzymes involved in glucose and lipid metabolism.
One of the challenges in drug development is achieving specificity, as many enzymes have similar active sites. However, advances in structural biology and computational modeling have led to the development of more specific inhibitors that target allosteric sites or other unique features of enzymes.
Final words on Enzyme Regulation
Enzyme regulation is a fundamental aspect of cellular metabolism, ensuring that biochemical pathways operate efficiently and respond appropriately to the cell’s needs. Through mechanisms such as allosteric regulation, covalent modification, proteolytic cleavage, compartmentalization, and gene expression, cells can finely tune enzyme activity to maintain homeostasis.
Dysregulation of enzyme activity can lead to a variety of diseases, including cancer, metabolic disorders, and neurological disorders. Understanding the mechanisms of enzyme regulation is therefore crucial for the development of new therapies that target these diseases.
As research in this field continues to advance, we can expect to gain new insights into the complex interplay between enzymes and their regulators, paving the way for more effective treatments and a deeper understanding of the molecular basis of life.
Summary/Key Takeaways:
- Enzyme regulation is essential for maintaining cellular homeostasis and adapting to changing conditions.
- Multiple mechanisms exist to control enzyme activity, including allosteric regulation, feedback inhibition, and covalent modification.
- Regulation can occur at various levels, from rapid changes in enzyme conformation to long-term alterations in gene expression.
- Understanding enzyme regulation is crucial for comprehending cellular metabolism and developing therapeutic interventions for metabolic disorders.
The study of enzyme regulation serves as a testament to the elegant complexity of biological systems and the power of evolution to craft sophisticated solutions to the challenges of life. As we continue to unravel the mysteries of enzyme regulation, we move closer to a comprehensive understanding of the intricate dance of molecules that underlies all living processes.
Frequently Asked Questions
What is the difference between competitive and non-competitive inhibition in enzyme regulation?
Competitive inhibition occurs when an inhibitor molecule binds to the enzyme’s active site, directly competing with the substrate. Non-competitive inhibition involves the inhibitor binding to a site other than the active site, causing a conformational change that reduces enzyme activity. Competitive inhibition can be overcome by increasing substrate concentration, while non-competitive inhibition cannot.
How does phosphorylation regulate enzyme activity?
Phosphorylation is a covalent modification that adds a phosphate group to specific amino acid residues (usually serine, threonine, or tyrosine) on an enzyme. This modification can activate or inhibit the enzyme by inducing conformational changes, altering substrate binding, or affecting interactions with other molecules. Phosphorylation is reversible, allowing for the dynamic regulation of enzyme activity in response to cellular signals.
What role does enzyme regulation play in metabolic diseases?
Enzyme regulation is crucial for maintaining proper metabolic function. Disruptions in regulatory mechanisms can lead to various metabolic diseases. For example, mutations affecting the regulation of enzymes involved in glucose metabolism can contribute to diabetes. Understanding enzyme regulation is essential for developing treatments for metabolic disorders and identifying potential therapeutic targets.
How do allosteric effectors differ from substrates in enzyme regulation?
Allosteric effectors bind to a site on the enzyme distinct from the active site (the allosteric site), while substrates bind to the active site. Allosteric effectors modulate enzyme activity by inducing conformational changes, affecting the enzyme’s affinity for its substrate or its catalytic efficiency. Substrates, on the other hand, are the molecules directly involved in the chemical reaction catalyzed by the enzyme.
Can environmental factors influence enzyme regulation?
Yes, environmental factors can significantly impact enzyme regulation. Temperature, pH, and the presence of certain ions or molecules can affect enzyme structure and function, influencing their regulatory properties. Additionally, environmental signals can trigger cellular responses that lead to changes in enzyme regulation through mechanisms such as altered gene expression or post-translational modifications.