The Ramachandran plot, named after the Indian physicist G.N. Ramachandran, is a fundamental concept in structural biology and biochemistry. This powerful visualization tool provides crucial insights into protein structure and has become an indispensable part of protein research and drug design. In this comprehensive article, we will delve deep into the world of Ramachandran plots, exploring their history, significance, and applications in modern science.
Table of Contents
What is a Ramachandran Plot?
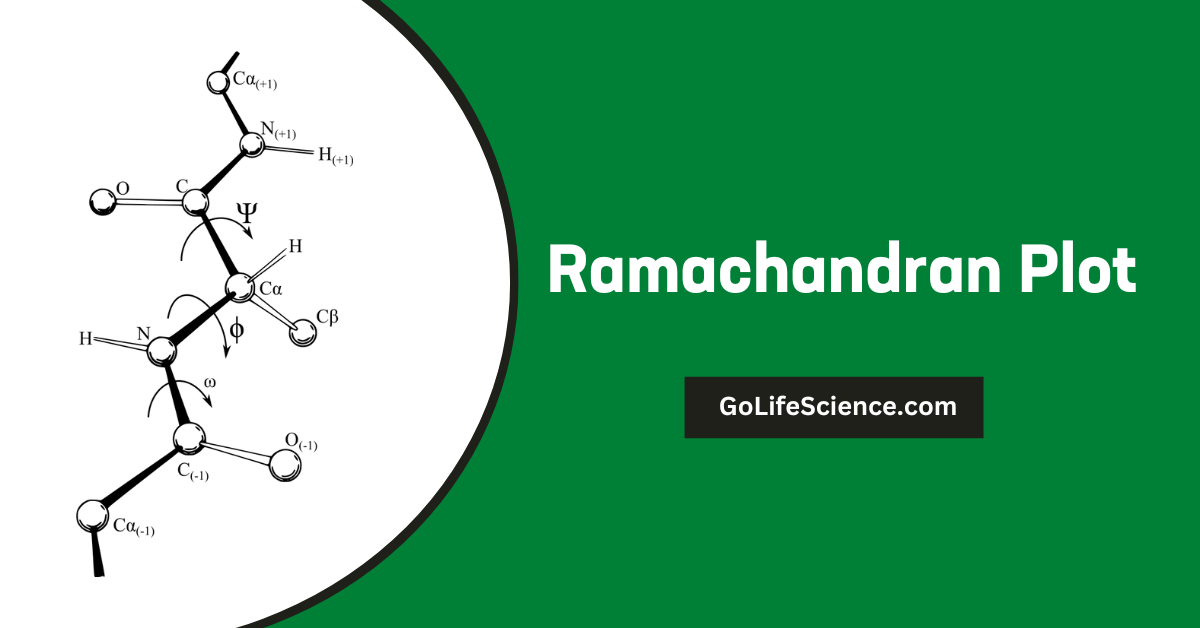
A Ramachandran plot, also known as a Ramachandran diagram or a φ-ψ plot, is a way to visualize energetically allowed regions for backbone dihedral angles ψ (psi) against φ (phi) of amino acid residues in protein structures. These plots serve as a powerful tool for assessing the quality of protein structures and understanding the conformational preferences of polypeptide chains.
The Backbone Dihedral Angles: Phi (φ) and Psi (ψ)
To understand Ramachandran plots, we first need to grasp the concept of backbone dihedral angles:
- Phi (φ) angle: This is the angle of rotation around the N-Cα bond.
- Psi (ψ) angle: This is the angle of rotation around the Cα-C bond.
These angles determine the three-dimensional structure of the protein backbone. The Ramachandran plot maps these two angles against each other, with φ on the x-axis and ψ on the y-axis.
Historical Background
The Ramachandran plot was first introduced in 1963 by G.N. Ramachandran, C. Ramakrishnan, and V. Sasisekharan in their groundbreaking paper titled “Stereochemistry of Polypeptide Chain Configurations.” This work laid the foundation for understanding the allowed conformations of polypeptide chains in proteins.
Ramachandran and his colleagues used basic principles of geometry and stereochemistry to calculate the allowed combinations of φ and ψ angles. They considered the steric clashes between atoms and identified regions in the plot where these angles could exist without causing unfavorable atomic overlaps.
Who is G. N. Ramachandran?
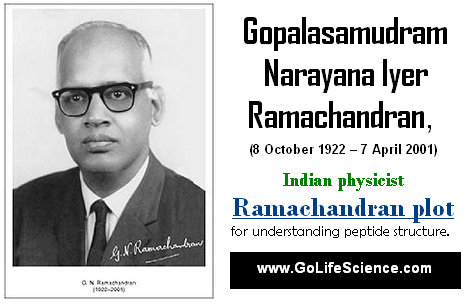
Gopalasamudram Narayana Ramachandran (8 October 1922 – 7 April 2001) is an Indian biophysicist and crystallographer who, along with Gopinath Kartha, worked out the triple helical structure of collathe basicgen.
G. N. Ramachandran is an Indian biophysicist who was known for his work that led to his creation of the Ramachandran plot for understanding peptide structure. He was the first to propose a triple-helical model for the structure of collagen. He also made other major contributions in biology and physics.
Features of Ramachandran Plot
- X-axis and Y-axis: The x-axis represents the phi (φ) angle, typically ranging from -180° to +180°. The y-axis represents the psi (ψ) angle, also ranging from -180° to +180°.
- Allowed Regions: These are areas in the plot where the φ and ψ angle combinations are energetically favorable. They are often colored or shaded to indicate different levels of favorability.
- Disallowed Regions: These are areas where φ and ψ angle combinations result in steric clashes between atoms. These regions are typically left blank or lightly shaded.
- Secondary Structure Regions: Specific areas of the plot correspond to common secondary structures like α-helices, β-sheets, and left-handed α-helices.
Interpreting the Plot
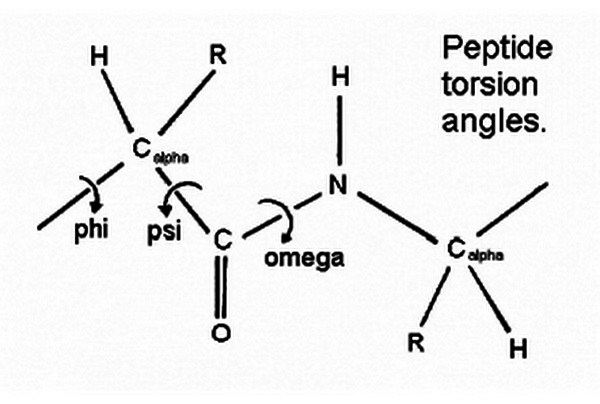
- Core Regions: The most favorable regions, often referred to as “core” regions, are typically found in the following areas:
- Right-handed α-helix: φ ≈ -60°, ψ ≈ -45°
- β-sheet: φ ≈ -120°, ψ ≈ 120°
- Left-handed α-helix: φ ≈ 60°, ψ ≈ 45°
- Allowed Regions: These are areas surrounding the core regions where conformations are still energetically favorable but less common.
- Generously Allowed Regions: These are regions where conformations are possible but less common and may indicate strained or unusual structures.
- Disallowed Regions: These are areas where conformations are highly unfavorable due to steric clashes.
Secondary structure plot regions
Secondary structures of a peptide are segments of the peptide that have an ordered and repetitive structure, and the repetitive structure is due to a repetitive confirmation of the residues and, ultimately, repetitive values of ψ and φ. The different secondary structures can be distinguished by their range of ψ and φ values, with the values of different secondary structures mapping to different regions of the Ramachandran plot. Two common examples of secondary structure are illustrated below.
α-helix
The scene on the right shows the axis of the α-helix rotating in the y-plane. When viewing the helix on the end, observe the open center of the helix. Planes are drawn on some of the peptide bonds to emphasize that in an α-helix, the planar peptide bonds rotate about the axis of the helix.
The Ramachandran plot of this peptide has points clustered about the values of ψ = -47o and φ = -57o, which are the average values for α-helices.
Adding the values of two other helical segments demonstrates that data from all three appear in one large cluster and that the helical segments cannot be distinguished by the differences in their ψ and φ values.
β-sheets
Display two-segment twisted β-sheets. Draw the planes of the peptide bonds. Most β-sheets in globular proteins are twisted sheets that do not have even parallel pleads.
Closer view of β-sheets. The Ramachandran plot of this twisted sheet has points clustered about the values of ψ = -135o and φ = +135o, which are the average values for twisted sheets.
Adding the values of three other sheet segments more clearly defines the area in which values for twisted sheets are located.
Plot Regions Limited by Steric Hindrance
The Ramachandran plot is fundamentally based on the concept of steric hindrance, which plays a crucial role in determining the allowed and disallowed regions. Let’s delve deeper into this aspect:
- Steric Forbidden Regions: Most combinations of ψ and φ angles are sterically forbidden due to atomic clashes. This can be illustrated using a tripeptide model, such as Glu-Ser-Ala. When the Serine residue adopts values of ψ = -116° and φ = 55°, its side chain comes into close contact with the Alanine residue, creating an unfavorable steric clash.
- Clustering in Allowed Regions: In Ramachandran plots of native peptides, data points form clusters in several areas where steric hindrance is minimal or absent. These clusters correspond to energetically favorable conformations.
- Core Regions: The core regions contain the most favorable combinations of ψ and φ angles and typically have the highest density of data points. These regions often correspond to common secondary structures like α-helices and β-sheets.
- Allowed Regions: The allowed regions (often shown in green in Ramachandran plots) surround the core regions or can be separate from them. They contain fewer data points than the core regions but still represent energetically favorable conformations.
- Generous Regions: These extend beyond the allowed regions and represent conformations that are less common but still possible in certain contexts.
- Disallowed Regions: The remaining areas of the plot are considered disallowed regions. Data points falling in these areas often indicate potential issues with the protein structure or unusual functional constraints.
Example of Steric Effects
Let’s consider our Glu-Ser-Ala tripeptide example:
- When Serine has ψ = -116° and φ = 55°, it falls in the disallowed region in the lower right quadrant of the Ramachandran plot. This conformation leads to steric clashes.
- However, if the Serine adopts ψ = -47° and φ = -57°, which falls in the core region in the lower left quadrant, its side chain rotates away from the Alanine, avoiding steric clashes.
This example illustrates how the Ramachandran plot effectively captures the conformational preferences dictated by steric constraints.
Special Case: Glycine
Glycine deserves special attention in the context of Ramachandran plots:
- Unique Flexibility: Since glycine has only a hydrogen atom as its side chain, it experiences much less steric hindrance compared to other amino acids. This allows glycine to adopt a wider range of ψ and φ angle combinations.
- Expanded Allowed Regions: In a Ramachandran plot, glycine residues often appear in regions that would be disallowed for other amino acids. For example, in a tripeptide Glu-Gly-Ala, the glycine can adopt ψ and φ values of -116° and +55° respectively without causing the steric hindrance observed in the Glu-Ser-Ala example.
- Identification in Plots: In many Ramachandran plots, a significant portion of the data points found in traditionally disallowed regions are often glycine residues.
Functionally Relevant Residues
While the Ramachandran plot is an excellent tool for assessing protein structure quality, it’s important to note that some functionally critical residues may adopt unusual conformations:
- Energetically Unfavorable but Functionally Important: Some residues crucial for a protein’s function may have torsion angles that plot in the disallowed regions of a Ramachandran plot.
- Catalytic Sites: This is particularly common in enzyme active sites, where the specific geometry required for catalysis may force residues into energetically unfavorable conformations.
- Structural Uniqueness: These unusual conformations, while rare, can be essential for the protein’s specific function or structural integrity.
- Careful Interpretation Needed: When analyzing Ramachandran plots, residues in disallowed regions shouldn’t be automatically flagged as errors. Instead, they warrant careful examination to determine if they serve a specific functional role.
By understanding these nuances in Ramachandran plot interpretation, researchers can gain deeper insights into protein structure-function relationships and more accurately assess the quality of protein structure models.
How to Read and Interpret a Ramachandran Plot
A Ramachandran plot is a powerful visualization tool that maps the backbone dihedral angles ψ (psi) against φ (phi) of amino acid residues in protein structures. Understanding how to read and interpret these plots is crucial for protein structure analysis and validation.
1. Dual Purpose of Ramachandran Plots
Ramachandran plots serve two primary purposes:
- Theoretical Conformations: They illustrate which values or conformations of the ψ and φ angles are theoretically possible for an amino acid residue in a protein. This aspect is based on stereochemical constraints and helps in understanding the fundamental principles of protein structure.
- Empirical Distribution: They display the actual distribution of data points observed in a single protein structure or a set of structures. This usage is particularly valuable for structure validation and quality assessment.
2. Key Regions in a Ramachandran Plot
The Ramachandran plot is typically divided into several regions, each representing different levels of conformational favorability:
- Core Regions: These are the darkest areas on the plot, representing the most favorable combinations of phi-psi values. In high-quality structures, over 90% of the residues should ideally fall within these core regions. The percentage of residues in the core regions is considered one of the best indicators of a structure’s stereochemical quality.
- Allowed Regions: These areas surround the core regions and represent phi-psi combinations that are less common but still energetically favorable.
- Generous Regions: Introduced by Morris et al. (1992), these regions extend approximately 20° (two 10° x 10° pixels) around the allowed regions. Residues falling in these areas are relatively rare and warrant closer investigation.
- Disallowed Regions: These are areas where phi-psi combinations result in steric clashes between atoms. Residues found in these regions often indicate potential errors in the structure, unless they serve a specific functional role.
3. Ramachandran Plot Analysis
When analyzing a Ramachandran plot, consider the following:
- Distribution of Data Points: Examine how the data points are distributed across the plot. A high-quality structure will have most points clustered in the core and allowed regions.
- Outliers: Pay special attention to any points falling in the generous or disallowed regions. While these may indicate structural issues, they can also highlight functionally important residues.
- Secondary Structure Patterns: Different secondary structures cluster in specific areas of the plot. For example, α-helices typically cluster around φ = -60°, ψ = -45°, while β-sheets cluster around φ = -120°, ψ = 120°.
- Glycine Residues: Remember that glycine residues have more conformational freedom and may appear in regions that would be disallowed for other amino acids.
- Resolution Dependence: The quality of the Ramachandran plot can be influenced by the resolution of the protein structure. Higher resolution structures generally show tighter clustering in the favored regions.
4. Quantitative Analysis
Modern Ramachandran plot analysis often includes quantitative measures:
- Percentage in Core Regions: As mentioned earlier, having over 90% of residues in the core regions is a good indicator of high structural quality.
- Ramachandran Z-score: This score compares the distribution of phi-psi angles in a structure to the expected distribution based on high-quality reference structures.
- Residue-specific Analysis: Some tools provide residue-specific Ramachandran plots, allowing for more detailed analysis of individual amino acids or regions of interest.
5. Historical Context and Modern Developments
The regions defined in Ramachandran plots have evolved over time:
- Original Studies: The original Ramachandran plot was based on theoretical calculations and limited structural data available in the 1960s.
- Empirical Refinement: The regions described by Morris et al. (1992) were based on observed phi-psi distributions from 121,870 residues in 463 known X-ray protein structures, providing a more accurate representation of favored conformations.
- Modern Datasets: Contemporary Ramachandran plot analysis often uses even larger datasets of high-resolution structures, leading to more refined definitions of the allowed regions.
- Resolution-specific Plots: Some modern tools offer resolution-specific Ramachandran plots, accounting for the fact that the spread of allowed conformations can vary with structure resolution.
6. Beyond Static Analysis
While traditional Ramachandran plots provide valuable insights into static structures, modern structural biology is increasingly concerned with protein dynamics:
- Ensemble Analysis: For NMR structures or molecular dynamics simulations, Ramachandran plots can be generated for entire ensembles, providing insights into conformational flexibility.
- Time-resolved Plots: With advances in time-resolved structural biology techniques, there’s growing interest in developing methods to visualize how Ramachandran distributions change over time during protein function or folding.
- Integration with Other Data: Modern structural biology often integrates Ramachandran plot analysis with other types of data, such as sequence conservation, B-factors, or functional annotations, to provide a more comprehensive understanding of protein structure-function relationships.
By mastering the interpretation of Ramachandran plots, researchers can gain deep insights into protein structure, validate experimental models, guide structure refinement, and even inform protein design efforts. As our understanding of protein structure continues to evolve, so too will the ways we use and interpret these invaluable plots.
Applications of Ramachandran Plots
Ramachandran plots have numerous applications in structural biology and related fields:
1. Protein Structure Validation
One of the primary uses of Ramachandran plots is to validate protein structures determined by experimental methods such as X-ray crystallography or NMR spectroscopy. By plotting the φ and ψ angles of all residues in a protein structure, researchers can quickly identify any residues that fall into disallowed regions, indicating potential errors in the structure determination or refinement process.
2. Protein Structure Prediction
In the field of protein structure prediction, Ramachandran plots serve as a valuable guide. They help in assessing the quality of predicted structures and can be used to refine computational models by ensuring that the backbone conformations adhere to the allowed regions of the plot.
3. Protein Engineering and Design
When designing new proteins or modifying existing ones, scientists use Ramachandran plots to ensure that the engineered sequences will adopt the desired conformations. This is particularly important in the development of novel enzymes, therapeutics, and biomaterials.
4. Understanding Protein Folding
Ramachandran plots provide insights into the energetics and conformational preferences of polypeptide chains, contributing to our understanding of protein folding mechanisms. They help explain why certain secondary structures are more common than others and how local interactions influence the overall fold of a protein.
5. Analyzing Protein Dynamics
By generating Ramachandran plots from molecular dynamics simulations, researchers can study the conformational flexibility of proteins. This approach helps in understanding protein function, ligand binding, and allosteric mechanisms.
6. Identifying Unique Structural Features
Ramachandran plots can reveal unusual or rare conformations in proteins, which may be linked to specific functional or structural roles. For example, certain regions of the plot are associated with tight turns or loops that may be critical for protein function.
Advanced Concepts in Ramachandran Plots
1. Non-Glycine vs. Glycine Plots
Glycine, the simplest amino acid with only a hydrogen atom as its side chain, has much more conformational flexibility compared to other amino acids. As a result, separate Ramachandran plots are often used for glycine residues:
- Non-Glycine Plot: This is the standard Ramachandran plot used for all amino acids except glycine. It shows more restricted allowed regions due to the presence of side chains.
- Glycine Plot: This plot shows a much larger allowed region, reflecting the greater conformational freedom of glycine residues.
2. Pre-Proline Ramachandran Plots
Proline residues have a unique cyclic structure that restricts their conformational freedom. Moreover, the residue preceding a proline (pre-proline) also has distinct φ and ψ angle preferences. Pre-proline Ramachandran plots are used to analyze these specific conformations.
3. High-Resolution Ramachandran Plots
With the increasing availability of high-resolution protein structures, more detailed Ramachandran plots have been developed. These plots provide finer gradations of allowed regions and can reveal subtle preferences in backbone conformations that were not apparent in earlier, lower-resolution plots.
Ramachandran Plots in the Era of Big Data
The exponential growth of the Protein Data Bank (PDB) has led to new insights and applications of Ramachandran plots:
1. Statistical Analysis
Large-scale analysis of φ and ψ angles from thousands of protein structures has led to more accurate definitions of allowed regions and a better understanding of the frequency distribution of different conformations.
2. Machine Learning Applications
Machine learning algorithms are being applied to Ramachandran plot data to improve protein structure prediction, identify novel conformations, and enhance our understanding of sequence-structure relationships.
3. Integration with Other Data
Researchers are combining Ramachandran plot data with other types of structural and functional information to gain more comprehensive insights into protein behavior and evolution.
Limitations and Considerations
While Ramachandran plots are incredibly useful, it’s important to be aware of their limitations:
- Side Chain Effects: Ramachandran plots primarily focus on backbone conformations and do not directly account for side chain interactions, which can significantly influence local structure.
- Context Dependency: The allowed regions can vary depending on the local environment of a residue within the protein structure.
- Dynamic Nature of Proteins: Ramachandran plots represent static conformations, while proteins are dynamic entities in solution.
- Resolution Dependence: The quality and interpretation of Ramachandran plots can be affected by the resolution of the protein structure data.
Future Directions
The field of structural biology continues to evolve, and with it, the applications and interpretations of Ramachandran plots:
- Integration with Cryo-EM Data: As cryo-electron microscopy (cryo-EM) becomes more prevalent in structure determination, new approaches to applying and interpreting Ramachandran plots for these structures are being developed.
- Time-Resolved Plots: With advancements in time-resolved structural biology techniques, there’s potential for developing dynamic Ramachandran plots that capture conformational changes over time.
- Artificial Intelligence and Deep Learning: AI and deep learning approaches are being explored to extract more nuanced information from Ramachandran plots and to predict protein structures with higher accuracy.
- Incorporation of Quantum Mechanics: More sophisticated energy calculations, including quantum mechanical effects, may lead to refined definitions of allowed regions in Ramachandran plots.
Conclusion
The Ramachandran plot, conceived over half a century ago, remains a cornerstone of structural biology. Its elegance lies in its simplicity – reducing the complex three-dimensional structure of proteins to a two-dimensional plot of two critical angles. From validating experimental structures to guiding protein design and unraveling the mysteries of protein folding, Ramachandran plots continue to be an indispensable tool in the structural biologist’s arsenal.
As we move forward into an era of big data, artificial intelligence, and increasingly sophisticated experimental techniques, the Ramachandran plot is likely to evolve and find new applications. However, its fundamental principle – providing a clear, visual representation of the conformational preferences of polypeptide chains – will undoubtedly continue to illuminate our understanding of protein structure and function for years to come.
Frequently Asked Questions (FAQs) on Ramachandran Plot
What is the main purpose of a Ramachandran plot?
The main purpose of a Ramachandran plot is to visualize the distribution of backbone dihedral angles (φ and ψ) in protein structures. It serves as a powerful tool for assessing the quality of protein structures, understanding conformational preferences of polypeptide chains, and validating experimentally determined or computationally predicted protein structures.
Why are some regions in the Ramachandran plot more favored than others?
Certain regions in the Ramachandran plot are more favored due to the stereochemical constraints of the polypeptide chain. These regions correspond to energetically stable conformations where there are minimal steric clashes between atoms. The most favored regions typically correspond to common secondary structures like α-helices and β-sheets, which are stabilized by hydrogen bonding patterns.
How does the Ramachandran plot differ for glycine compared to other amino acids?
Glycine, being the smallest amino acid with only a hydrogen atom as its side chain, has much more conformational flexibility than other amino acids. As a result, the Ramachandran plot for glycine shows a much larger allowed region compared to the plots for other amino acids. This is because glycine can adopt φ and ψ angle combinations that would cause steric clashes in other amino acids due to their larger side chains.
Can Ramachandran plots be used to identify errors in protein structures?
Yes, Ramachandran plots are frequently used to identify potential errors in protein structures. If a significant number of residues fall into disallowed regions of the plot, it may indicate problems with the structure determination or refinement process. However, it’s important to note that some residues may legitimately adopt unusual conformations due to functional constraints or local environmental factors.
How are Ramachandran plots used in protein structure prediction?
In protein structure prediction, Ramachandran plots serve as a guide for assessing the quality of predicted structures and for refining computational models. They are often used as part of the scoring function in structure prediction algorithms, penalizing conformations that fall into disallowed regions. Additionally, the statistical preferences observed in Ramachandran plots from known structures can be used to inform the sampling of conformations during the prediction process.